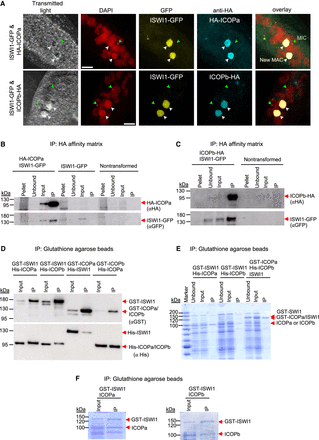
Interaction of ICOPa and ICOPb with ISWI1 in new MACs. (A) Confocal fluorescence microscopy images of HA-ICOPa, ICOPb-HA, and ISWI1-GFP localization: maximum intensity projections of z-planes. Samples were collected when ∼90% of cells had visible anlagen. Red indicates DAPI; yellow, GFP; cyan, HA; green arrow, MIC; and white arrow, new MAC. All channels were optimized individually for the best visual representation. DAPI channel of ICOPb-HA: Gamma factor = 0.8. Scale bar = 10 µm. (B,C) Western blot; coimmunoprecipitation (co-IP) of HA-ICOPa/ISWI1-GFP and ICOPb-HA/ISWI1-GFP in Paramecium. Controls are nontransformed and ISWI1-GFP-transformed. HA-ICOPa/ICOPb-HA: 94 KDa; ISWI1-GFP: 147 KDa. (D–F) Co-IP after E. coli expression and pulldown. (D) Western blot; (E,F) Coomassie staining. (D–F) GST-ISWI: 147 kDa; His-ISWI1: 122 kDa; His-ICOPa and His-ICOPb: 95 kDa; GST-ICOPa/ICOPb: 119 kDa; untagged ISWI1: 120 kDa; and untagged ICOPa and ICOPb: 93 kDa.











