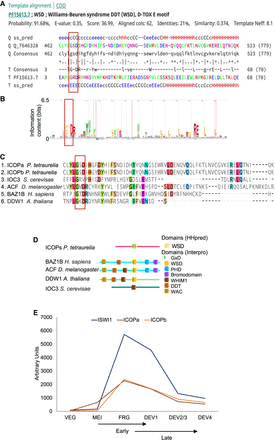
Identification of ISWI complex proteins (ICOPs). (A) Template alignment generated by HHpred analysis of ICOPa showing a 91.68% probability match (E-value = 0.35) with Williams–Beuren syndrome DDT (WSD) or D-TOX E motif. The conserved GxD signature is highlighted with a red bar. (Q) Query (ICOPa), (ss_pred) secondary structure prediction, and (T) template. For a more detailed output format description, please consult https://toolkit.tuebingen.mpg.de/tools/hhpred. (B) Signature of Pfam model PF15613 from InterPro. (C) Multiple sequence alignment of ICOPa and ICOPb WSD motif with WSD motif–containing protein regions from other organisms, including ISWI complex. (Ioc3) ISWI one complex protein 3 in yeast, (ACF) ATP-dependent chromatin assembly factor large subunit (Acf) from D. melanogaster, (BAZ1B) human tyrosine-protein kinase BAZ1B, and (DDW1) DDT domain-containing protein in A. thaliana. Red box indicates GxD signature; highlighted amino acids with ≥50% of residues identical to the consensus residue. (D) Domain architecture comparison of ICOPs with ISWI1 complex proteins with WSD motifs. (E) mRNA expression profile (arbitrary units) of ICOPa and ICOPb in comparison to ISWI1 during autogamy based on data from ParameciumDB (Arnaiz et al. 2020). (VEG) vegetative, (MEI) the stage at which MICs undergo meiosis and maternal macronucleus (MAC) begins to fragment, (FRG) ∼50% of cells with fragmented maternal MAC, (DEV1) the earliest stage with visible developing MACs (anlage), (DEV2/3) most cells with macronuclear anlage, and (DEV4) most cells with distinct anlage. In this paper, we consider “early” development to be MEI and FRG (T6–T10) and “late” development to be DEV1 and DEV2/3 (T12–T16).











