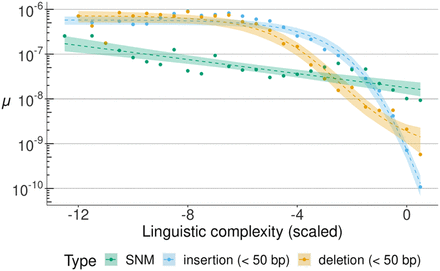
Relationship between SNM and indel rates (on a log10 scale) and sequence repetitiveness. Sequence repetitiveness was measured as the linguistic complexity of 101 bp genomic windows and then scaled to standard deviation units, with a mean of zero. On this scale, more negative values indicate more repetitive sequences. The observed rates of different types of mutations (in colors) are shown in data points. Dashed lines represent regression model fits of mutation rate against linguistic complexity. For SNMs, a linear model was fitted, whereas indel data fitted best a logistic regression; 95% confidence intervals are presented for all regression models.











