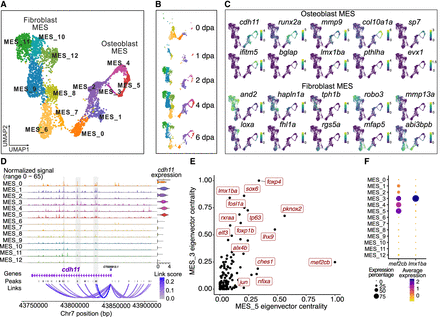
MES sublineages and lineage-specific regulatory networks. (A) A UMAP plot shows MES subclusters. Cells are colored by subclusters. (B) A set of UMAP plots divided from A by time points. (C) A set of UMAP plots with embedded RNA expression of marker genes. (D) A genome browser view generated with Signac CoveragePlot displays pseudobulk snATAC-seq signals for each subcluster around the cdh11 gene. Annotations at the bottom indicate the transcript region, ATAC-seq peaks, and peak-to-gene links. Violin plot on the right side shows the normalized RNA expression levels of cdh11 across each cluster. Tracks and violin plots are colored by subclusters. (E) A scatter plot shows the top TFs in the MES_3 and MES_5 subclusters, ranked by their eigenvector centrality scores. (F) A dot plot showing RNA expression of mef2cb and lmx1ba across subclusters.











