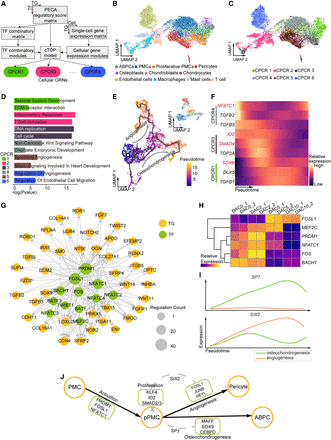
cTOP models cell programs in antler regeneration. (A) cTOP couples cell expression modules and TF combinatory modules under the PECA regulatory network to model cellular regulatory programs. (B,C) UMAP of cell types and highest CPCR in cell atlas at dac5. (D) Functional enrichment for TGs of each CPCR. (E) UMAP of pseudotime trajectory of stromal cells at dac5 by Monocle3. (F) cTOP-relative TFs (red) and genes (black) expression dynamics across the osteochondrogenesis trajectory. (G) Network of CPCR4 suggests PRDM1, FOSL1, and NFATC1 as the key factors for stem cell activation in antler regeneration. (H) Expression dynamics of TFs in CPCR4 during antler regeneration suggest their early function in antler regeneration. (I) Expression dynamic across differentiation of SP7 and SIX2 showing their program specific expression. (J) Schematic regulatory model of cellular programs in antler stem cell differentiation. Black arrows represent differentiation, and gray dashed arrows represent regulation.











