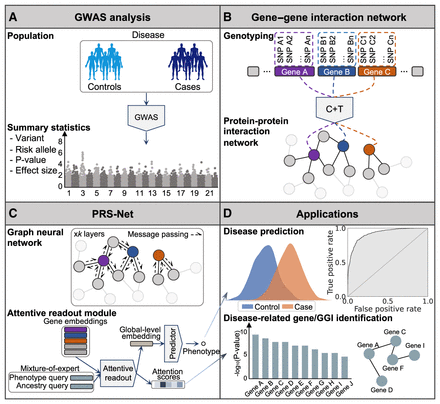
An illustrative diagram of PRS-Net. (A) The proposed framework is built upon GWAS summary statistics, including variants, risk alleles, P-values, and effect sizes. (B) A gene–gene interaction (GGI) network is constructed based on the protein–protein interactions (PPIs) in this study. Gene-level PRSs (various P-value thresholds applied) are calculated using the C + T method, serving as the node features within the network. (C) A graph neural network (GNN) is employed to update node features via message passing, and subsequently, an attentive readout module is introduced to facilitate model interpretation. (D) PRS-Net can be applied for both disease prediction and gene discovery. (GWAS) genome-wide association stuty, (C + T) clumping and thresholding.











