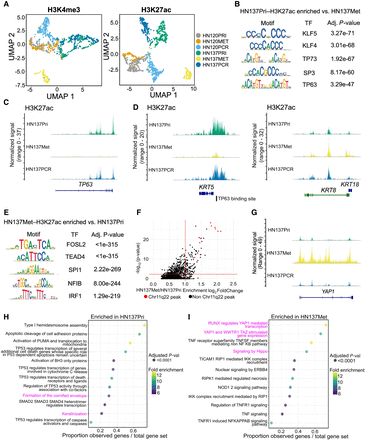
Epigenomic changes during HNSCC progression suggest distinct, patient-specific epigenetic drivers of tumor evolution. (A) UMAP embedding for H3K4me3 (left) and H3K27ac (right) for HN120 and HN137 single-cells. (B) Enriched transcription factor (TF) motifs for HN137Pri derived from H3K27ac peaks. (C) Coverage plot of H3K27ac signal at the TP63 locus, showing loss of H3K27ac in HN137Met. (D) Coverage plots of H3K27ac signal at the KRT5 (left) and KRT8/18 (right) loci. TP63 binding site near the KRT5 promoter is indicated. (E) Enriched TF motifs for HN137Met. (F) Top enriched H3K27ac peaks in HN137Met, with peaks at the Chr11q22 locus containing YAP1 highlighted in red. (G) Coverage plot of H3K27ac signal at the YAP1 locus. (H,I) Dot plot indicating the REACTOME pathways enriched in HN137Pri (H) and HN137Met (I). Highlighted in magenta are enriched terms relating to differentiation in HN137Pri- and YAP1-mediated loss of differentiation in HN137Met.











