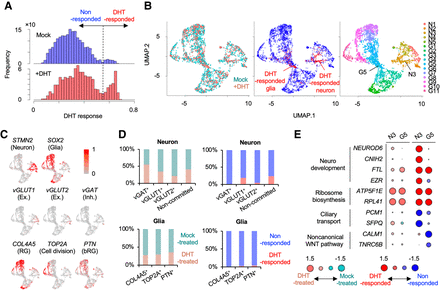
Inference of response to DHT treatment in brain organoids by weakly supervised deep learning. (A) Histograms of DHT response score in DHT- and mock-treated organoids. Dashed line is a threshold to define DHT-responsive cells (DHT response score > 0.55). (B) UMAP plot of single cells colored by organoid type (left), DHT response (middle), and cluster (right): (N) neuron, (G) glia. (C) UMAP plot showing representative gene expression for each cell type. (D) Ratio of cells from DHT-treated and mock-treated organoids (left) and DHT- and nonresponded cells (right) in each cell type. (E) Representative differentially expressed genes in N3 and G5 cluster between DHT- and mock-treated cells (left) and between DHT- and nonresponded cells (right). Circle size represent log2(ratio).











