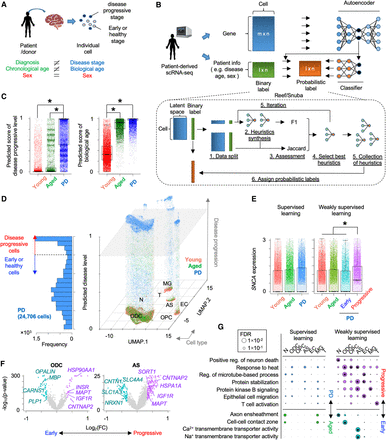
Inference of Parkinson's disease (PD) progression levels of individual cells from patient-derived scRNA-seq. (A) Difference between patient information and status of individual cells in patient-derived tissue. (B) Schematic of weakly supervised deep learning model to infer disease-progressed cells. (C) Boxplots showing predicted score of disease progressive level (left) and biological age (right). (*) P < 0.05 by two-sided t-test. (D) 3D plot showing disease-progressed cells in each cell type. The x- and y-axes represent the first and second dimensions of UMAP. The z-axis is the predicted disease progression level. Histogram of predicted disease level in PD patient–derived cells is shown in left panel. The dashed line and gray plane represent threshold of disease progression and early-staged/healthy cells. (E) Comparison of synuclein alpha (SNCA) expression across groups that are segregated by supervised (left) or weakly supervised (right) deep learning. (F) Volcano plots showing differential expression between PD progressive and early-staged/healthy cells. (G) Circle plot showing overrepresented GO terms between PD progressive and early-staged/healthy cells (right) but not between PD and aged brain (left). Circle size represents –log10(FDR). Statistical significance (FDR < 0.05) is shown by an asterisk. (N) Neuron, (ODC) oligodendrocyte, (OPC) oligodendrocyte precursor cell, (AS) astrocyte, (EC) endothelial cell, (MG) microglia, and (T) T cell.











