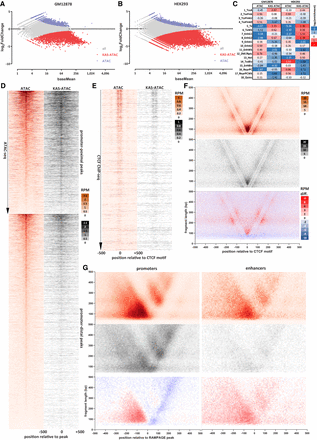
The single-stranded accessible chromatin landscape is distinct from total open chromatin genomic maps. (A) DESeq2 analysis of GM12878 ATAC and KAS-ATAC libraries. (B) DESeq2 analysis of HEK293 ATAC and KAS-ATAC libraries. (C) Enrichment of KAS-ATAC/ATAC differential peaks over ChromHMM chromatin states. Enrichment is calculated as the log2 ratio of the observed versus expected fractions of peaks overlapping each state (based on the overlap of the total set of peaks with ChromHMM states). (D) ATAC-seq and KAS-ATAC profiles around promoter-proximal and distal peaks (GM12878 cells). (E) ATAC-seq and KAS-ATAC profiles around occupied CTCF sites in GM12878 cells (excluding promoter-proximal CTCF peaks; CTCF ChIP-seq peak calls were obtained from ENCODE accession ENCFF827JRI). (F) ATAC-seq, KAS-ATAC, and KAS-ATAC/ATAC differential V-plot (Henikoff et al. 2011) around occupied CTCF sites. (G) ATAC-seq, KAS-ATAC, and KAS-ATAC/ATAC differential V-plot around RAMPAGE (Batut et al. 2013) peaks in promoter and in distal CRE regions (RAMPAGE peaks were obtained from ENCODE accession ID ENCSR000AEI).











