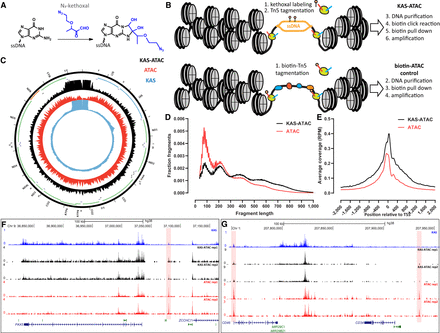
Overview of the KAS-ATAC assay. (A) N3-kethoxal specifically covalently labels unpaired guanine bases. (B) KAS-ATAC captures DNA fragments that are simultaneously single-stranded and accessible, by first labeling ssDNA with N3-kethoxal and then carrying out an ATAC-seq reaction. After purification of DNA and a click reaction that conjugates biotin onto kethoxal-labeled DNA, accessible ssDNA is specifically pulled down using streptavidin beads and amplified. In parallel, a control “biotin-ATAC” library can be prepared using biotinylated Tn5 that then undergoes the same pull-down procedure as KAS-ATAC DNA fragments (if the goal is to quantify the relative abundances of accessible ssDNA and accessible DNA). (C) KAS-seq, ATAC-seq, and KAS-ATAC mitochondrial genome profiles in human GM12878 cells. (D) Fragment length distribution in biotin-ATAC-seq and KAS-ATAC libraries (GM12878 cells). (E) Genome-wide TSS metaprofiles for biotin-ATAC-seq and KAS-ATAC libraries (GM12878 cells). (F,G) Representative genome browser snapshots showing ATAC-seq, KAS-seq, and KAS-ATAC profiles around nuclear loci in GM12878 cells.











