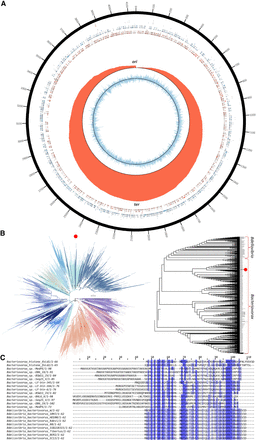
Genome assembly and annotation of Bacteriovorax sp. ICPB 3264 [H-I A3.12]. (A) Circos (Krzywinski et al. 2009) plot of the Bacteriovorax sp. ICPB 3264 [H-I A3.12] chromosome. Forward- and reverse-strand protein-coding genes are shown in red and blue, respectively, in the inner tile layer; ribosomal RNAs and tRNAs are shown in the outer tile layers. The red histogram shows the cumulative GC skew (Grigoriev 1998), with the inflection points corresponding to the replication origins and termini (Lobry 1996). The light blue histogram in the innermost circle shows the GC skew for each individual bin along the genome. (B) Place of Bdellovibrionota in the global prokaryote phlyogeny (obtained from Zhu et al. 2019 at https://biocore.github.io/wol/gallery/ranks/order.pdf) on the left and the maximum likelihood 16S rRNA tree (generated using RAxML-NG) (Kozlov et al. 2019) of members of the Bdellovibrionota phylum and the phylogenetic location of strain ICPB 3264 [H-I A3.12] on the right. Bdellovibrionota is highlighted with a red dot on the left, and the strain we worked with is highlighted with a red dot on the right. (C) Multiple sequence alignment of histone proteins identified in select Bacteriovorax and Bdellovibrio genomes. In contrast to the singlet histone folds in Bdellovibrio, the histone gene in Bacteriovorax sp. ICPB 3264 [H-I A3.12] is a histone doublet; the two histone folds are shown separately (top two rows).











