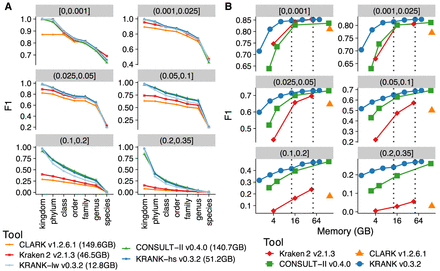
Detailed evaluation of read classification performances of tools across different ranks and memory levels. (A) Comparison of two default modes of KRANK (lw for lightweight and hs for high-sensitivity) with other methods across ranks and novelty bins on the WoL-v1 data set with 66,667 simulated 150 bp reads from each of 756 query genomes. Each panel is a novelty bin, corresponding to the minimum distance to any reference genome (d*), measured by Mash (Ondov et al. 2016); see Supplemental Figure S1. F1 scores are averages of all queries in each d* bin. See Supplemental Figure S3 for precision and recall. (B) F1 accuracy for KRANK, CONSULT-II, and Kraken 2 as we change the memory used in GB. CLARK is shown as a single data point. We mark memory levels corresponding to the lw and hs modes using dashed lines.











