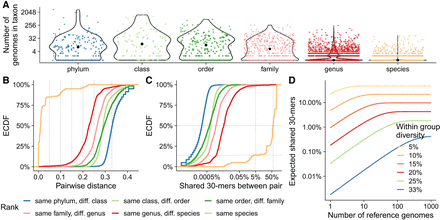
Exploratory analysis of imbalanced taxon sampling and the portion of shared k-mers across taxonomic ranks in the WoL-v1 data set (Zhu et al. 2019). (A) Number of reference genomes under each taxonomic node (dots), separated by ranks. (B,C) The distribution of Mash (Ondov et al. 2016) estimated genomic distances (B) and Jaccard similarities (C) among 500,000 randomly sampled pairs of genomes that share a taxonomic rank but are different in lower ranks. The empirical cumulative distribution function (ECDF) is shown. (D) The theoretical expectation for the number of 30-mers shared between a query and at least one of N sampled genomes of a reference set for a group that has within-group diversity 2d is shown as (1 − d)k(1 − (1 − (1 − d)k)N).











