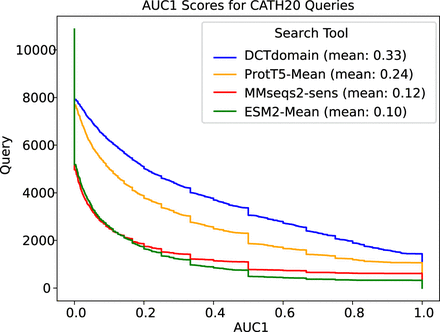
AUC1 plots for comparison of the different methods on the CATH20 database search benchmark that contains distant homologs. As reported in their paper (Schütze et al. 2022), using ProtT5 mean embeddings (i.e., knnProtT5) outperforms MMseqs2-sensitive. DCTdomain outperforms ProtT5-Mean by a similar margin, whereas using the mean embedding method with ESM-2 performs slightly worse than MMseqs2-sens (sensitive mode).











