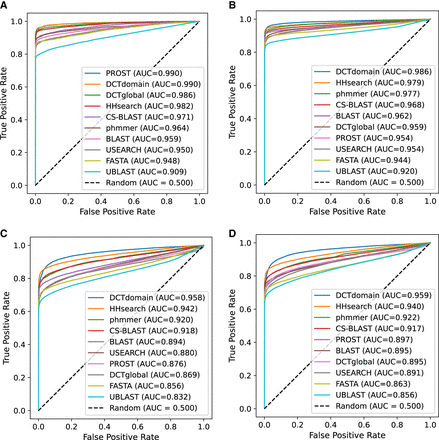
ROC plots for comparison of the different methods on four benchmarks of global homologs. (A) pfam-max50 benchmark; (B) pfam-nomax50 benchmark; (C) gene3d-nomax50 benchmark; and (D) supfam-nomax50 benchmark. We replicated the AUC values found in Saripella et al. (2016) for popular homology detection tools, as well as adding results for most similar DCT domain fingerprints (DCTdomain) and similarity between global fingerprints (DCTglobal) between protein pairs. We find that the DCTdomain performs the best on every benchmark, with a higher separation between tools on the gene3d and supfam data sets, which take domains from structural-based databases.











