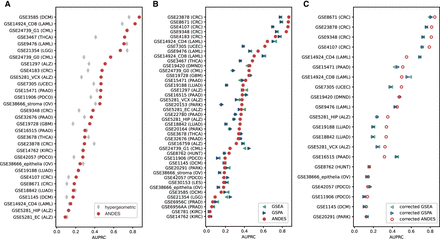
ANDES achieves state-of-the-art performance in overrepresentation-based and rank-based gene set enrichment for the GEO2KEGG (Tarca et al. 2013) gene set enrichment benchmark. (A) Performance comparison between ANDES and hypergeometric test in retrieving annotated KEGG terms using genes that have FDR ≤ 0.05 in each data set (where there are at least 10 genes that are significantly differentially expressed). (B) Performance comparisons between ANDES, GSEA (Subramanian et al. 2005), and GSPA (Cousins et al. 2023) in retrieving annotated KEGG terms using the full list of genes (no FDR cutoff), ranked by log2(fold change). In both cases, ANDES statistically outperforms other methods, demonstrating the advantage of incorporating gene embedding information using the best-match principle into the gene set enrichment setting. (C) Performance comparisons between ANDES, GSEA, and GSPA with empirically estimated P-values in retrieving annotated KEGG terms. Only expression data sets with at least 10 samples in both normal and diseased conditions (21 data sets) are included for sufficient variability in label permutations. Corrected-ANDES still outperforms corrected-GSEA and corrected-GSPA.











