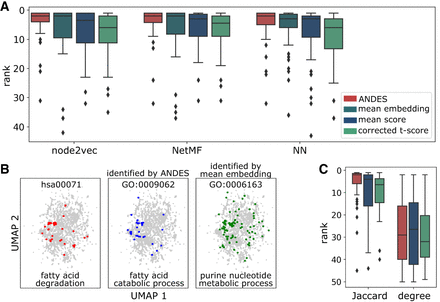
ANDES better matches gene sets that describe the same biological processes regardless of the underlying embedding or network structure. (A) Boxplots of the ranking of the correct matching GO term for 50 KEGG terms demonstrate that ANDES outperforms the mean embedding, corrected t-score, and mean score methods across three network embedding approaches (node2vec, NetMF, NN). NN is a structure-aware autoencoder method (Methods). We also note that the handful of KEGG–GO pairs where ANDES performs poorly have consistently poor performance across methods (e.g., none of the five ANDES outlier KEGG terms in node2vec achieve a better ranking in any other methods). (B) UMAP of the node2vec PPI network embedding of genes in the KEGG fatty acid degradation gene set highlights a failure of the mean embedding method to capture meaningful substructure. Inspection of the embedding space reveals a similar substructure between the correct KEGG–GO term match prioritized by ANDES that is not seen in the top match for the mean embedding method. (C) Baseline approach for gene set matching in PPI networks. Matched KEGG–GO terms are ranked using pairwise similarity based on gene neighbor Jaccard similarity (Jaccard), or more naively, by the sum of node degrees (degree). Because these pairwise similarity matrices are directly calculated from network properties without using embeddings, we cannot calculate the mean embedding method and instead compare ANDES to only the corrected t-score and mean score.











