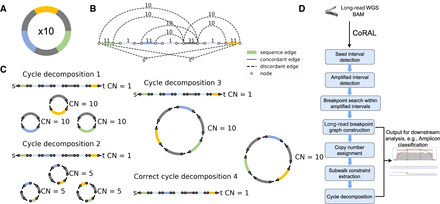
Long-read-based ecDNA reconstruction. (A) Native ecDNA structure and copy number. (B) Cartoon of the breakpoint graph derived from the ecDNA in A. Sequence edges represent segments of the reference genome. Concordant edges connect consecutive sequences with respect to the reference genome order, and discordant edges connect nonconsecutive genome segments. Nodes are created at the endpoints of each sequence edge and include source and sink nodes, s and t. (C) Multiple collections of decomposed paths and cycles from the breakpoint graph explain the changes in copy number and observed SVs. Long reads that span regions of high multiplicity can help resolve the correct cycle. (D) Overview of the CoRAL method.











