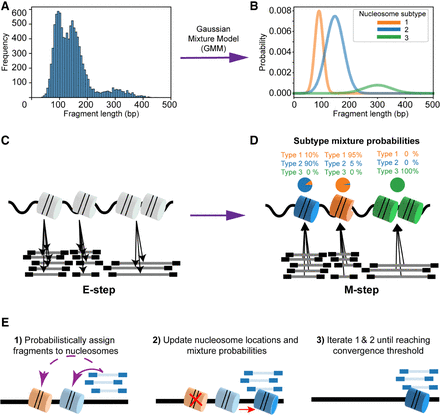
Overview of the SEM algorithm. (A) SEM first calculates the global fragment size distribution from the MNase-seq data. (B) A Gaussian mixture model (GMM) is used to deconvolve the fragment size distribution into a set of nucleosome subtypes. (C) In the expectation step of the algorithm, each MNase-seq fragment is probabilistically assigned to nucleosome components according to the current locations, strengths, and subtype identities of the components. (D) In the maximization step of the algorithm, the various nucleosome properties are updated based on the current fragment assignments. (E) Detailed illustration of how nucleosome properties are updated during EM iterations.











