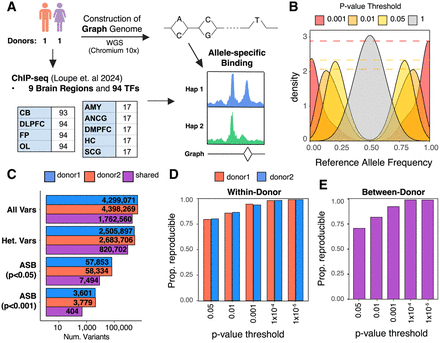
Overview of experimental design and allele-specific binding (ASB). (A) Workflow for detection of ASB. Whole-genome sequencing and ChIP-seq of 94 TFs were performed in postmortem brain samples from two donors. ChIP-seq reads were mapped to personalized graph genomes and tested for ASB across donors and tissues. Abbreviations denote the following: (DLPFC) dorsolateral prefrontal cortex, (FP) frontal pole, (OL) occipital lobe, (CB) cerebellum, (AnCg) anterior cingulate, (SCg) subgenual cingulate, (DMPFC) dorsomedial prefrontal cortex, (Amy) amygdala, and (HC) hippocampus. (B) Density plot showing the distribution of reference allele frequency at heterozygous variants for different P-value thresholds. (C) The number of total phased variants, heterozygous variants, significant variants (P < 0.05), and significant variants (P < 0.001) for both donors. “Shared” indicates variants that are shared and pass filters in both donors. (D) Within-donor reproducibility. The fraction of ASB events that preferred the same allele across brain regions at increasing P-value threshold. (E) Between-donor reproducibility. The fraction of ASB events that favored the same allele when comparing the same TF–allele interaction between donors.











