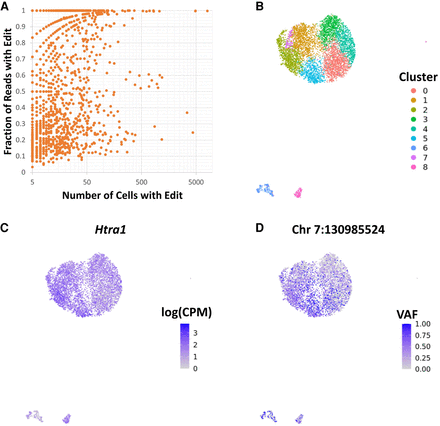
Evidence of potential RNA-editing events in scRNA-seq variants, demonstrated by (A) a high incidence of editing sites and the presence in both low and high portions of reads. (B–D) UMAP embedding of cells in EPSRC1 scRNA-seq sample based on per-cell expression profile, with clusters derived from Louvain clustering of normalized log counts per million for each gene after regressing cell-cycle effects to demonstrate the heterogeneity of cells (B); expression level of the gene on which the RNA edit of interest is located (LOG(CPM) indicates logarithm of counts per million), demonstrating a uniform expression profile (C); RNA-edit allele presentation frequency (VAF) differing across cells in a pattern similar to expression-based cell heterogeneity (D).











