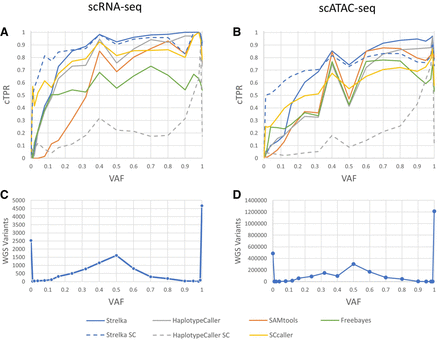
Figure 1.
Common true-positive rate (cTPR) averaged over samples for different variant-calling approaches, for scRNA-seq (A) and scATAC-seq (B), binned by VAF (e.g., the point VAF = 0.5 would include variants from 0.5 to the next bin, which is 0.6). The number of WGS variants with coverage in the single-cell sequencing modality, for scRNA-seq (C) and scATAC-seq (D), binned by VAF.











