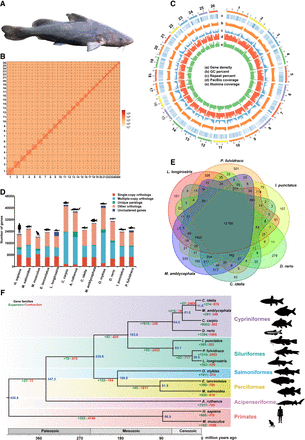
Chromosome-level genome assembly of the Chinese longsnout catfish (Leiocassis longirostris Günther) and the comparative genomics analysis with 10 species from mammals to teleosts. (A) Representative photo of Chinese longsnout catfish. (B) Genome-wide chromosomal contact map of Chinese longsnout catfish based on chromatin interaction data sets analyzed with Hi-C. Among chromosomes, a heat map from yellow to red intensity was used to represent the Hi-C links interaction frequency distribution (yellow to red represents low to high intensity). (C) Visualization of the whole-genome alignment is shown using the Circos plot (Krzywinski et al. 2009). Outer to the inner circles: chromosome length (each tick mark represents 1/5 Mb), gene density, GC percentage distribution (in nonoverlapping 100 kb windows), repeat percentage, Pacific Biosciences (PacBio) coverage, and Illumina coverage. (D) Distribution of genes of different types in different species. Single-copy genes and multiple-copy genes are the gene families found in the 13 species. Single-copy genes correspond to the number of genes in a gene family in which only one species has genes. Other genes refer to the number of genes in gene families that are not species specific. The number of nonclustered genes in each species. (E) Venn diagram showing the overlap of orthologous genes in Chinese longsnout catfish and five other fishes, including yellow catfish (Pelteobagrus fulvidraco), channel catfish (Ictalurus punctatus), zebrafish (Danio rerio), grass carp (Ctenopharyngodon idellus), and blunt snout bream (Megalobrama amblycephala). (F) Phylogenetic tree and time tree showing the expansion and contraction of gene families in 13 species ranging from mammals to teleosts, namely, grass carp (C. idellus), blunt snout bream (M. amblycephala), common carp (Cyprinus carpio), zebrafish (D. rerio), channel catfish (I. punctatus), yellow catfish (P. fulvidraco), Chinese longsnout catfish (L. longirostris Günther), rainbow trout (Oncorhynchus mykiss), giant grouper (Epinephelus lanceolatus), largemouth bass (Micropterus salmoides), sterlet (Acipenser ruthenus), Homo sapiens, and mouse (Mus musculus). The blue numbers indicate the times of divergence between the different species; the green numbers indicate the number of gene families that have expanded over the course of evolution; and the red numbers indicate the numbers of gene families that have contracted.











