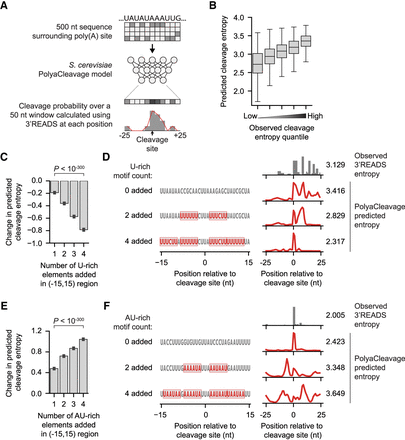
Cleavage heterogeneity can be modulated by changing the poly(A) site surrounding cis-regulatory elements. (A) Workflow describing our design of the S. cerevisiae PolyaCleavage model. (B) Box plots showing the increase in predicted cleavage entropy as the observed cleavage entropy measured by 3′READS increases. Cleavage sites from the holdout testing set were split into five evenly sized groups (N ∼ 809 sites per group). (C) The change in predicted cleavage entropy as nonoverlapping U-rich elements are sequentially introduced in the [–15,+15] region surrounding the maximum cleavage site. The data are presented as the mean and the 95% confidence interval (error bar). The P-value from the Wilcoxon signed-rank paired test comparing the addition of one U-rich element to four U-rich elements is shown. Consensus high entropy sites with flanking UA-rich motifs and no surrounding U-rich elements were included (N = 182 actual cleavage sites, 29,056 altered sequences). (D) An example showing the effect of introducing U-rich elements around the cleavage site in a high entropy cleavage site from YIL156W-B. The predicted entropy values are shown. (E) The change in predicted cleavage entropy as nonoverlapping AU-rich elements are sequentially introduced in the [–15,+15] region immediately surrounding the maximum cleavage site. The data are presented as the mean and the 95% confidence interval (error bar). The P-value from the Wilcoxon signed-rank paired test comparing the addition of one AU-rich element to four AU-rich elements is shown. Consensus low entropy sites with flanking U-rich motifs were included (N = 222 actual cleavage sites, 35,012 altered sequences). (F) Similar to D, showing the effect of introducing AU-rich elements around the cleavage site in a low entropy poly(A) site from YPR154W.











