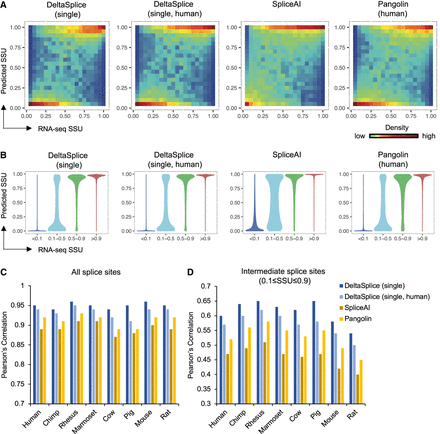
The performance of DeltaSplice in single-sequence mode and baseline methods in predicting SSU across different species. (A) Distribution of human SSU predicted by different methods in comparison with experimental measurements by RNA-seq. DeltaSplice (single) refers to DeltaSplice in single-sequence mode trained with data from all species, and DeltaSplice (single, human) was the model in single-sequence mode trained using only human data. Pangolin was retrained with human SSU data to facilitate a fair comparision, considering that the original Pangolin model was trained with SSU estimated from different RNA-seq data set using a different implementation. The SpliceAI model from the original study, which was trained on human gene splice sites, was used here. (B) Similar to A, but splice sites are binned into four groups based on RNA-seq SSU values. The four groups have 83,251 (<0.1), 6370 (0.1–0.5), 10,672 (0.5–0.9), and 113,752 (>0.9) splice sites, respectively. (C) Pearson's correlation between RNA-seq and predicted SSU for all splice sites. (D) Similar to B, but for splice sites with experimental SSU between 0.1 and 0.9.











