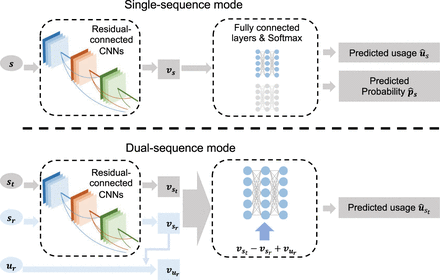
The architecture of DeltaSplice. DeltaSplice comprises a feature-extraction module and multiple prediction modules. The feature-extraction
module is constructed using residual-connected convolutional neural networks (CNNs), which convert a one-hot-encoded input
sequence to a feature representation. Each prediction module consists of fully connected layers and a Softmax output layer
that takes a feature representation as input and generates predictions for SSU or splice-site probabilities. The single-sequence
mode employs two prediction modules to predict the SSU 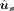 and the splice-site probabilities
and the splice-site probabilities 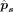 for each site in the input gene sequence s, based on the corresponding feature representation vs. In the dual-sequence mode, the feature-extraction module calculates the feature representation
for each site in the input gene sequence s, based on the corresponding feature representation vs. In the dual-sequence mode, the feature-extraction module calculates the feature representation 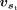 and
and 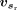 separately for the target gene sequence st and the reference gene sequence sr. The predicted SSU
separately for the target gene sequence st and the reference gene sequence sr. The predicted SSU 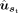 for every site in the target gene sequence is computed using a prediction module, from the input
for every site in the target gene sequence is computed using a prediction module, from the input 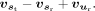 Here
Here 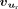 is the feature representation of the reference SSU ur. RNA-seq data from adult brain tissues of humans and seven other mammalian species, as summarized in Supplemental Table S1, were used to estimate SSU values for model training.
is the feature representation of the reference SSU ur. RNA-seq data from adult brain tissues of humans and seven other mammalian species, as summarized in Supplemental Table S1, were used to estimate SSU values for model training.











