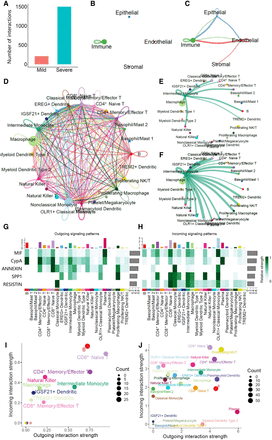
Comprehensive analysis of the cell communication network for COVID-19. (A) The number of interactions of the cell communication networks. (B,C) Cell communication networks at tissue compartment level for mild and severe patients, respectively. (D) Cell communication network of immune cell types based on integrated data. (E,F) Visualization of signaling pathways sending from intermediate monocyte cells for mild (E) and severe (F). Circle sizes are proportional to the number of cells in each cell type, and edge width represents the communication number. Edge colors are consistent with the source cell type. (G,H) The outgoing or incoming heatmap of each cell type to the inferred signaling pathways. The color bar represents relative signaling strength of a signaling pathway across cell types. The top colored bar plot shows the total signaling strength of a cell type by summarizing all signaling pathways displayed in the heatmap. The right gray bar plot shows the total signaling strength of a signaling pathway by summarizing all cell types displayed in the heatmap. (I,J) Total incoming and outgoing signaling strengths in different cell types for mild (I) and severe (J). The size of points matches the count of interactions.











