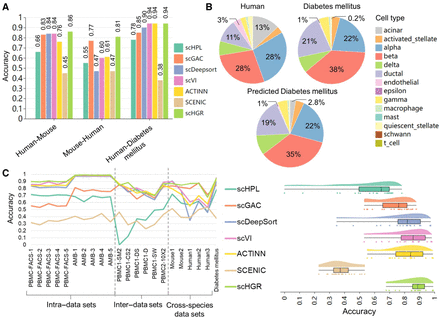
Performance comparison on multiple scenarios and summary of the overall experiments. (A) The bar chart exhibits the accuracy of all the approaches in three scenarios, which include annotating mouse cells by referring to human data sets (human–mouse) and vice versa (mouse–human) and annotating diabetes mellitus human cells by referring to human data sets (human–diabetes mellitus). The specific values are marked above the corresponding bars. (B) For human and diabetes mellitus, pie charts are produced based on their a priori labels and the percentage of each type. In contrast, the predicted diabetes mellitus is drawn based on labels annotated by scHGR. Comparing human and diabetes mellitus, five types with the highest ratio variation are marked with specific values at corresponding parts. (C) A total of 22 scenarios are divided into three groups: intra–data sets, inter–data sets, and cross-species data sets. The line chart reflects the variation in accuracy. Note that scHPL identifies all cells in PBMC1-SM2 as root, so it fails to annotate their types. The right chart contains an accuracy-based scatter plot (each point corresponds to a scene); a box plot that summarizes the maximal, minimal, and median scores, etc.; and a cloud plot that illustrates the distribution of accuracy values across various data sets.











