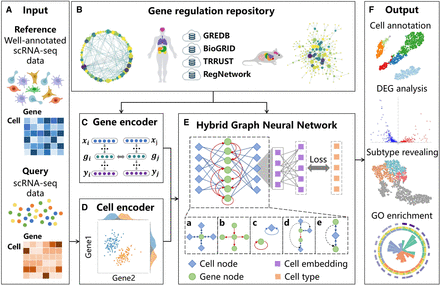
Overview of the scHGR. (A) The input of scHGR includes reference data sets equipped with single-cell resolution gene expression profiles as well as type information for each cell, and query data sets equipped with scRNA-seq data. (B) The underlying gene regulatory relationships to train scHGR are mainly from GREDB, BioGRID, TRRUST, and RegNetwork, which contain more than 2 million records pertaining to humans and mice. (C) Gene encoder generates the initial embedding of gene nodes via graph embedding algorithm. (D) Cell encoder encodes high-dimensional expression matrix into low-dimensional embedding of cell nodes. (E) A hybrid graph neural network (HGNN) factors in six types of edges. It optimizes parameters by minimizing the gap between prediction and reference cell types. The blue diamond and green circle represent the cell nodes and gene nodes in HGNN; the purple square represents cell embedding; and the orange square represents the cell types provided by reference data set. (F) The output of scHGR includes automatic cell annotation and differential expression gene (DEG) analysis based on propagation weight, revealing novel cell subtypes with biological significance and Gene Ontology (GO) enrichment.











