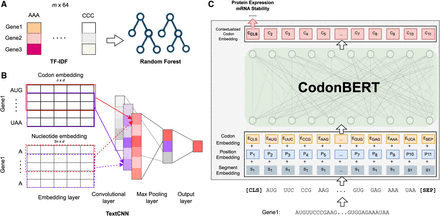
Comparison to prior methods (TF-IDF, Codon2vec, RNABERT, and RNA-FM) and fine-tuning CodonBERT on downstream data sets. (A) Given an input corpus with m mRNA sequences, TF-IDF is used to construct a feature matrix followed by a random forest regression model. (B) Use a TextCNN model to learn task-specific nucleotide or codon representations. The model is able to fine-tune pretrained representations by initializing the embedding layers with stacked codon or nucleotide embeddings extracted from pretrained language models (Codon2vec, RNABERT, and RNA-FM). n is the number of codons in the input sequence, and d is the dimension of the token embedding. As a baseline, plain TextCNN initializes the embedding layer with a standard normal distribution. (C) Fine-tune the pretrained CodonBERT model on a given downstream task directly by keeping all the parameters trainable.











