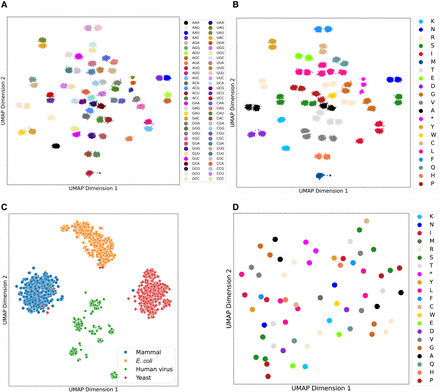
Genetic code and evolutionary taxonomy information learned by the pretrained, unsupervised CodonBERT model. High-dimensional embeddings were projected into two-dimensional space using UMAP (McInnes et al. 2018). (A,B) Projected codon embeddings from the pretrained CodonBERT model. Each point represents a codon with different contexts, and its color corresponds to the type of codon (A) or amino acid (B) accordingly. (C) Projected sequence embedding from the pretrained CodonBERT model. Each point is a mRNA sequence, and its color represents the sequence label. (D) Projected codon embedding from the pretrained Codon2vec model. Each point shows a codon, and its color is the corresponding amino acid.











