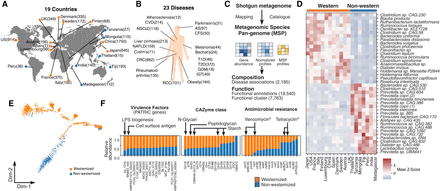
Characterization of the global gut microbiome in health and disease. Pan-metagenomics association studies of health and disease. Corresponding data sets were publicly shared as a resource: the Human Gut Microbiome Atlas (HGMA). (A) The geographical distribution of the data sets used in this study (the number of the samples is shown in parentheses). (B) Disease data sets of shotgun metagenomics used in this study. (C) The workflow of the metagenomic species pan-genome (MSP) quantification together with functional characterization. We first constructed 1989 MSPs for gut microbiome by MSPminer based on co-abundant gene profiles, which give clues to identify gene cluster markers likely belonging to the same species. Next, all the short reads aligned to the IGC2 catalog and, subsequently, gene abundances were profiled, downsized, and normalized. Based on co-abundant gene markers from the given MSP, mean signals were used to estimate species abundance profiles. In total, 6014 shotgun metagenome samples were aligned against the gene catalog of the human gut microbiome and quantified at the level of MSP. (D) Heatmap showing the top 20 significantly overrepresented MSPs between western and nonwestern cohorts colored by mean species Z-score for each country against all countries. (E) Monocle ordination of the gut microbiome. Individual samples from nonwestern and western countries were colored blue and orange, respectively. (F) Difference in gene content between western and nonwestern enriched species. Those species gene content was annotated by those that were CAZymes, antimicrobial-resistance (AMR) genes, and virulence factors (PATRIC database) and summed across all species. Total number of each gene was normalized and plotted as a stacked bar plot to show regional overrepresentation (Methods).











