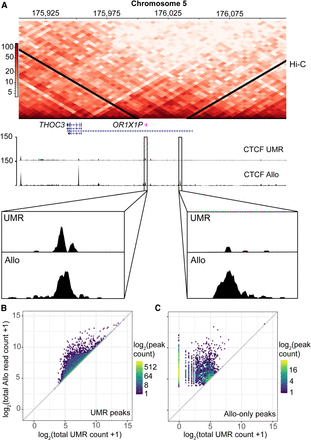
Allo increases the overall read counts at multimapped CTCF peaks and improves the resolution of uniquely mappable peaks. (A) Hi-C interaction heatmap showing two TAD boundaries on Chromosome 5 near the THOC3 gene. K562 CTCF read counts are also plotted for the UMR-only analysis and the Allo analysis. Bottom sections show the zoomed in version of two Allo-only peaks at these TAD boundaries. (B) Density scatterplot showing the total read count before and after the incorporation of Allo within CTCF UMR-derived peaks. (C) Density scatterplot showing the total read count before and after the incorporation of Allo within CTCF Allo-only peaks. Note that Allo was run using default settings, which allocates MMRs even in cases in which there are no nearby UMRs. This results in peaks called in regions with zero starting UMR counts as seen in panel C. The user can shut off this functionality using the argument “‐‐remove-zeros” to have more stringent criteria for MMR allocation.











