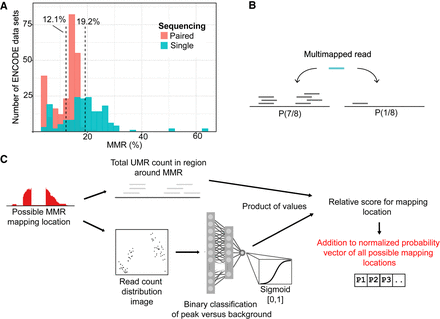
Figure 1.
Overview of the prevalence of multimapped reads (MMRs) in ChIP-seq data sets and Allo's algorithm for rescuing MMRs. (A) Proportions of reads that are multimapped across 481 K562 ENCODE TF ChIP-seq data sets. (B) Overview of probabilistic MMR allocation by proximal uniquely mapped read (UMR) count. (C) Overview of the Allo algorithm combining UMR counts and image classification of read distribution.











