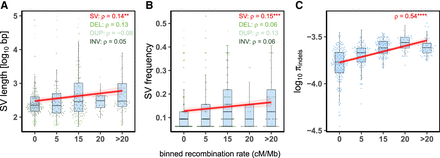
Association between recombination rates and genome dynamics in C. obscurior. Correlations between recombination rates and length (A) and frequency (B) of SVs and indel polymorphism (C) in 16 individual workers. The x-axis represents binned recombination rates, with the upper limit of each bin indicated. (ρ) Spearman's rank correlation coefficient is indicated for all structural variants (SVs) and separately for deletions (DEL), duplications (DUP), and inversions (INV), with significance shown by asterisks: (****) P < 0.0001, (***) P < 0.001, (**) P < 0.01. All correlations were performed using the raw data. Red lines and shaded areas are linear regressions and 95% confidence intervals.











