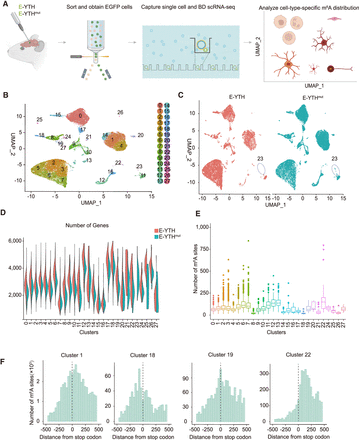
Hippocampal single-cell identification following AAV transduction. (A) Schematic diagram of detecting m6A RNA modification in the mouse hippocampus on a single-cell level through AAV transduction with E-Yth and E-Ythmut controls. (B) Integration of uniform manifold approximation and projection (UMAP) of 27,804 single-cell transcriptomes. Cluster numbers from zero to 27 are indicated. (C) Separate UMAP for E-YTH and E-YTHmut: 11,561 single cells in E-YTH and 16,243 in E-YTHmut. (Circle) Cluster 23 is missing in E-YTH. (D) Violin plot visualizing the number of genes per cluster. No genes were detected in cluster 23 in E-YTH samples. (Red) E-YTH, (blue) E-YTHmut. (E) m6A counts per cluster. (F) Metagene analysis of individual cell clusters identified by single-cell sequencing. The m6A number surrounding the stop codon (position 0) is shown. m6A sites were obtained after eliminating background editing sites. Clusters 1, 18, 19, and 22 are shown, which represent different m6A distribution patterns.











