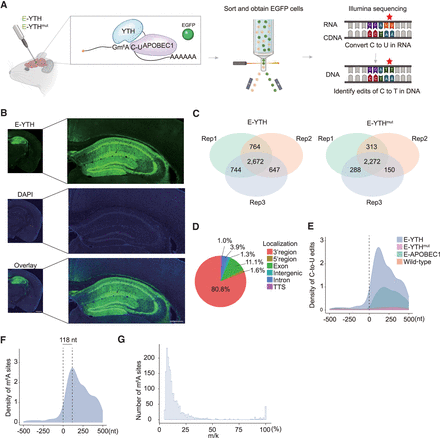
Detection of m6A with bulk RNA-seq in mouse hippocampus. (A) Schematic diagram of bulk RNA-seq in the mouse brain. The Egfp-Apobec1-Yth is packaged into AAV viruses to infect brain cells. EGFP-positive cells are isolated from the hippocampus and processed for C-to-U edit and m6A site identification. (B) Confocal image of mouse hippocampus after AAV infection. Representative image of E-YTH is shown. Half-brain image: Scale bar, 1 mm. Hippocampus image: Scale bar, 400 μm. (C) Number of overlapping C-to-U editing events identified by RNA-seq in hippocampus following Egfp-Apobec1-Yth and Egfp-Apobec1-Ythmut AAV virus injection and EGFP FACS sorting. Editing events identified in at least two replicates were considered for downstream analyses. n = 3, (Rep) Biological replicates from different animals. (D) Pie chart showing m6A localization identified by E-Yth in mouse hippocampus. (TTS) Transcription termination site. (E) Metagene analysis showing C-to-U edit scaled density 500 nt 5′ and 500 nt 3′ from stop codon (0 nt) in E-YTH, E-YTHmut, E-APOBEC1, and wild-type (WT) samples. The peak value for E-YTH is 2716 editing events. (F) Metagene analysis showing m6A density 500 nt 5′ and 500 nt 3′ from stop codon (0 nt). m6A sites were obtain after eliminating background from E-YTH editing sites. m6A peak density occurs 118 nt downstream from the stop codon. (G) Histogram of m6A site counts over mutation per read (m/k) ratio. Minimum threshold: 5%.











