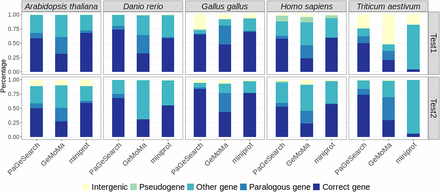
The percentage of annotations of regions identified in PaGeSearch, GeMoMa, and miniprot categorized by archetype species and the species used for validation. “Correct genes” are regions that overlap with the regions of the query genes; “paralogous genes” are regions that overlap with paralogs of the query genes; “other genes” are regions that overlap with protein-coding genes that are neither correct genes nor paralogs of the query genes; “pseudogenes” are regions that overlap with pseudogenes; and “intergenic” are regions that do not overlap with any genes.











