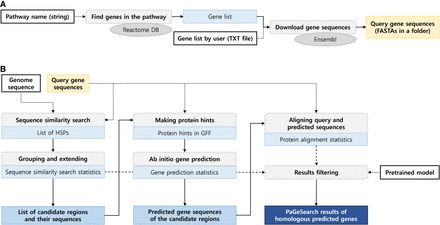
Process of PaGeSearch. (A) Genes in the specified pathway are identified from the Reactome database, and their sequences are downloaded from the Ensembl database for use as the PaGeSearch query. (B) PaGeSearch initiates by finding seed regions through a sequence similarity search, followed by gene prediction within these regions. The final results are filtered using a neural network model that evaluates sequence similarity, gene prediction metrics, and protein alignment statistics.











