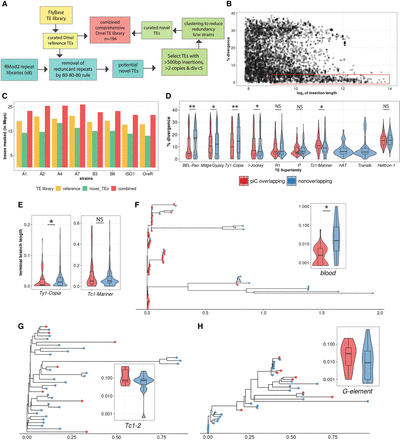
De novo TE annotation uncovers ∼3 Mb of hidden TEs and reveals strong associations of young LTR TEs with piCs more than any other TE subclasses. (A) TE annotation pipeline using RepeatModeler2 and RepeatMasker to create the comprehensive TE library. (B) Abundance of extremely similar and long TE insertions from RepeatMasker output of strain B6 using novel TE consensus library. (C) Differences in million base-pairs (Mbp) masked in RepeatMasker results using novel-only, reference-only, and combined TE library. (D) Divergence estimates for all defragmented iso-1 insertions (>200 bp) from RepeatMasker output. Insertions with >1-bp overlap with master-list iso-1 piCs are considered piC overlapping. Difference between groups is tested by Wilcoxon rank-sum test. (E) Terminal branch length for all iso-1 insertions from Ty1/Copia and Tc1/Mariner superfamilies from maximum likelihood trees. (F–H) Maximum likelihood trees constructed from all defragmented insertions for blood, Tc1-2, and G-element families, and the inset shows terminal branch length quantification. Difference between groups is tested by Wilcoxon rank-sum test: (*) P < 0.05, (**) P < 0.005, (***) P < 0.0005.











