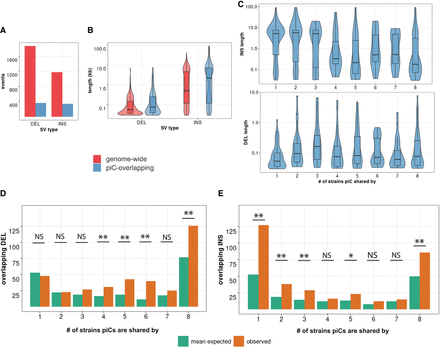
Common piCs show “trap” like sequence turnover. (A) Observed counts of indels genome-wide and overlapping with piCs. (B) Length distribution of indels genome-wide and overlapping with piCs. Significant differences are shown from Kruskal–Wallis test comparisons. (C) Length distribution of indels overlapping piCs grouped by the number of strains they are shared by (i.e., population frequency). (D,E) Enrichment analyses of deletion (DEL) and insertions (INS) variants in piCs performed. Variants overlapping with piCs of differing population frequencies are compared with the expected mean overlap counts genome-wide. Variant calls were shuffled 1000 times. (*) P < 0.05, (**) P < 0.005.











