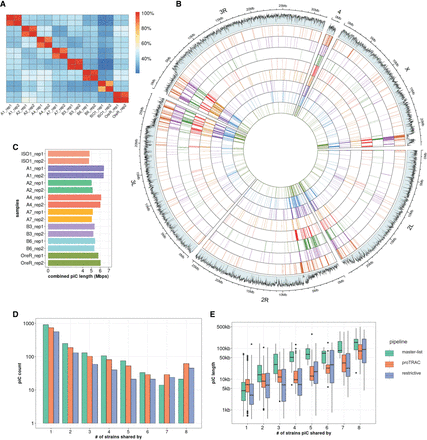
Interstrain variability of piCs in D. melanogaster strains. (A) Cross-strain and replicate overlap (percentage of total piCs count) of independently predicted piCs for each small RNA library using the restrictive method. Interstrain comparisons are performed after piCs were remapped to iso-1 coordinates, and overlap >1 bp between two piCs is deemed as shared. Intrastrain comparisons are performed similarly but in native genome assembly. (B) Genome-wide distribution of lifted-over piCs in seven DSPR strains and reference iso-1 strain. Bars along the circumference represent the presence of piCs in 10-kb bins for each chromosome. The outermost bar plot is piRNA mappability scores, followed by iso-1 piCs and then by piCs of seven DSPR strains. (C) Combined size of predicted piCs genome-wide from each replicate small RNA library independently in respective genome assemblies. (D) Population frequency of piCs quantified after liftover to the reference iso-1 genome. (E) piC length distribution by population frequency in kilobase pairs quantified after liftover to the iso-1 genome.











