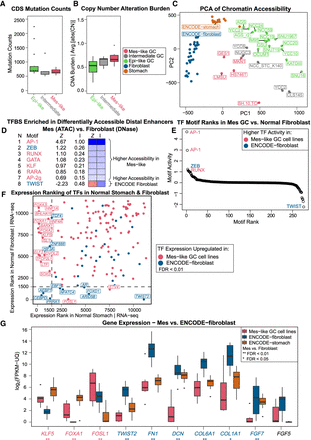
Despite some similarity in gene expression and enhancer activity, Mes-like GC and fibroblast have distinct TF responses. All groups of cell lines have similar levels of somatic mutations (A) and CN alterations (B). (C) PCA of chromatin accessibility of GC cell lines, stomach, and fibroblasts shows that although all groups are distinct, there is an axis along which differences in accessibility between stomach and fibroblasts are shared between the Epi-like and Mes-like cell lines. Epi-like cell lines are more similar to stomach, and Mes-like cell lines are more similar to but still distinct from, primary fibroblasts. (D) Mes-like versus fibroblast accessibility differences are driven by differential activity of a small set of TFs. (E) Rank plot of the average TF motif weights in all pairwise comparisons between Mes-like cell lines and ENCODE fibroblast samples. (F) Differential expression of TFs between the stomach and fibroblast shows that Mes-like cell lines retain high expression of stomach-specific TFs and do not upregulate all fibroblast TFs (especially TWIST2) to the same degree. Ranking of DE TFs between Mes-like and ENCODE-fibroblast (FDR < 0.01) in normal GTEx stomach (x-axis) and normal GTEx fibroblast profiles (y-axis) are shown. Some of the DE TFs upregulated in Mes-like GC (FOXA1, KLF5, etc.) have a higher expression (lower ranking) in GTEx normal stomach, whereas TWIST2 has a higher expression in ENCODE fibroblast and a higher expression (lower ranking) in GTEx fibroblast. (G) Inferred TF motif activities are supported by differential expression of stomach TFs and fibroblast genes in Mes-like GC, normal ENCODE-fibroblast, and ENCODE-stomach (**) FDR < 0.01, (*) FDR < 0.05, likelihood ratio test.











