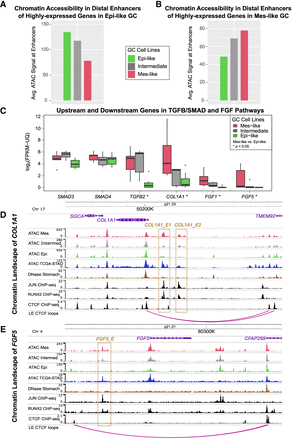
Mes and Epi upregulated genes are flanked by Mes-active and Epi-active distal enhancers. (A) Highly expressed genes in Epi-like GC cell lines are flanked by distal peaks with increased accessibility in Epi-like cell lines. (B) Highly expressed genes in Mes-like GC cell lines are flanked by distal peaks with increased accessibility in Mes-like cell lines. All peaks within 50 kb of TSS of DE genes were averaged. Many of the upregulated genes in Mes cell lines are members of the TGFB/SMAD and FGF pathways, and their expression is shown in C (Mann–Whitney U test P < 0.05). (D) In the COL1A1 locus, COL1A1_E1 and COL1A1_E2 are putative distal enhancers with increased ATAC-seq signal in Mes-like GC cell lines and TCGA-STAD relative to Epi-like and normal stomach, consistent with higher COL1A1 expression in Mes-like GC cell lines and TCGA STAD. ChIP-seq experiments in the mesenchymal liposarcoma cell line LPS141 identified AP-1 and RUNX2 binding in COL1A1_E1 and COL1A1_E2. (E) Similarly, in the FGF5 locus, FGF5_E is a putative distal enhancer with increased ATAC-seq signal in Mes-like GC cell lines and TCGA-STAD relative to Epi-like and normal stomach, consistent with higher FGF5 expression in Mes-like GC cell lines and TCGA STAD. FGF5_E is a RUNX2 and AP-1 binding site in LPS141. FGF5_E, COL1A1_E1, and COL1A1_E2 are contained in CTCF loops enclosing the target gene promoter.











