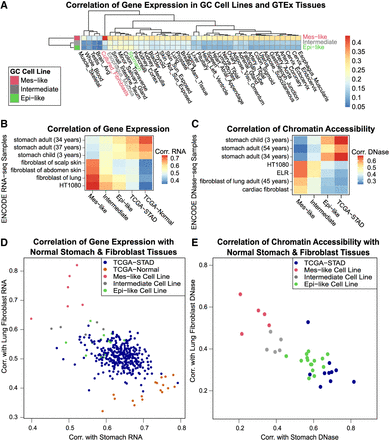
In both expression and chromatin accessibility, Mes-like GC cell lines are more similar to fibroblasts, and Epi-like GC cell lines are more similar to stomach. (A) Mes-like expression is most similar to GTEx fibroblasts. Correlation of 54 healthy GTEx gene expression profiles for each group of GC cell lines in about 11,300 tissue-specific genes. (B) Correlation of ENCODE stomach and fibroblast gene expression profiles with average gene expression for each group of GC cell lines. HT1080 is a fibrosarcoma cell line. (C) Mes-like chromatin accessibility is also most similar to an ENCODE fibroblast, and the TCGA-STAD and Epi-like GC cell lines are most similar to ENCODE stomach. We measured the similarity of chromatin accessibility by correlation of gkm-SVM weight vectors trained on distal enhancers of each sample. ELR is a fibroblast-derived cell line. (D) Correlation of GC cell line gene expression profiles with a healthy ENCODE lung fibroblast and a healthy stomach sample. (E) Correlation of GC cell line chromatin accessibility profiles (gkm-SVM weights trained on cell line ATAC-seq) with healthy ENCODE lung fibroblasts and stomach (gkm-SVM trained using DNase-seq).











