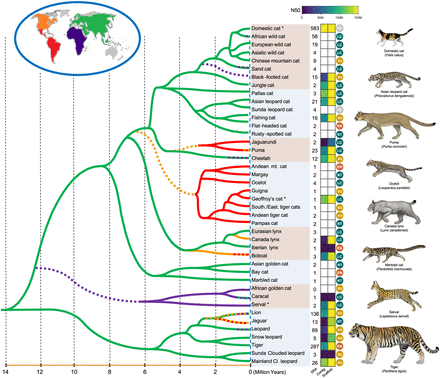
Phylogeny and evolutionary timescale of living felids. Branching relationships vary considerably across the genome, primarily as a function of gene flow/introgression and incomplete lineage sorting (Li et al. 2019). The relationships and species divergence times are derived from a consensus of regions of the genome with low recombination and of recent studies incorporating complete species sampling (Li et al. 2019; Jamieson et al. 2023; Lescroart et al. 2023; Yuan et al. 2023, 2024). The branches are color-coded based on current and inferred historical distributions from the fossil record and biogeographic reconstructions. Dashed branches indicate hypothesized dispersal events out of Europe and other regions. The columns shown to the right of the species names indicate the number of individuals with whole-genome sequence data sets available on NCBI's Sequence Read Archive (SRA) (last accessed January 11, 2024), as determined based on taxonomic classification (Kitchener et al. 2017; Lescroart et al. 2023). Heatmaps of contig (left) and scaffold (right) N50 metrics are shown from the highest-quality reference genomes from each respective species. Genomes nearing telomere-to-telomere (T2T) status are represented as assemblies with high contig and scaffold N50's (i.e., domestic cat), whereas species with low contig N50 but high scaffold N50 represent more fragmented assemblies. White boxes indicate species with no genome assembly available. IUCN conservation status logos are placed to the right of the assembly contiguity heatmaps. (NE) not evaluated, (LC) least concern, (VU) vulnerable, (EN) endangered, (NT) near threatened, (*) unpublished. Images by C. Buell.











