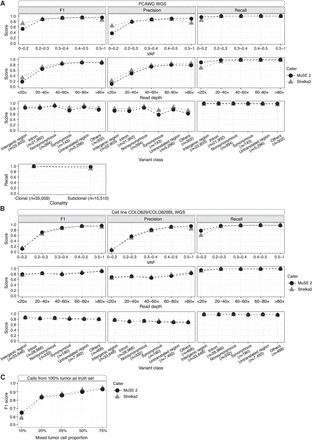
Accuracy benchmarking of MuSE 2 and Strelka2 within each bin of VAF (first row), sequencing read depth (second row), or class of variant effects (third row) for PCAWG WGS (A) and cell line WGS (B) data. Comparison of recall between the two methods within different clonality for PCAWG WGS data is shown in the last row of A. The calls of each method, as well as the truth set, are pooled from the WGS data of the selected five patient samples from PCAWG (A) or the WGS data of the cell line COLO829/COLO829BL generated from two platforms (B). The number of consensus calls for a variant class is included in the x-axis labels. (C) F1 scores of MuSE 2 and Strelka2 with varied tumor cell proportions. Calls from the tumor purity of 100% were used as the truth set.











