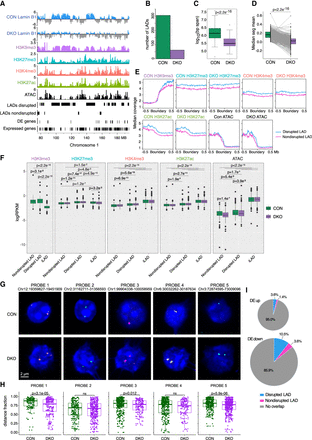
Suv39DKO reduces lamina-associated domains (LADs) without inducing gene activation. (A) Example tracks of Lamin B1 ChIP-seq from control and Suv39DKO DP thymocytes overlayed with H3K9me3, H3K27me3, H3K4me3, and H3K27ac ChIP-seq and ATAC-seq from control DP thymocytes. Disrupted LADs, not-disrupted LADS, DE genes, and expressed genes are also annotated. LADs are divided into LADs lost in DKO and LADs retained or gained. Lamin B1 ChIP-seq tracks are shown at 50-kb resolution, and all other tracks are shown at 10-kb resolution. (B) Number of LADs called by the enriched domain detector across all chromosomes in control and Suv39DKO DP thymocytes. (C) Distributions of LAD size from B in control and Suv39DKO DP thymocytes. Box plot depicts the IQR ± 1.5 × IQR and median annotated. Distributions were compared by Wilcoxon rank-sum test with continuity correction. (D) LAD strength score (seg.mean) from CBS algorithm in control and Suv39DKO DP thymocytes. Control and DKO scores were compared using a paired t-test. (E) Median coverage of H3K9me3, H3K27me3, H3K4me3, H3K27ac, and ATAC across LAD boundaries from CBS analysis showing disrupted LADs and nondisrupted LADs. (F) Distributions of H3K9me3, H3K27me3, H3K4me3, H3K27ac, and ATAC logRPKM in disrupted LADs (CBS score reduced by >0.1 in DKO vs. CON), not-disrupted LADs, and inter-LAD (iLAD) regions. Distributions are shown as box plots depicting the IQR ± 1.5 × IQR with median annotated. Distributions were compared by Wilcoxon rank-sum test with continuity correction. (G) DNA-FISH against LAD or iLAD loci in control and Suv39DKO DP thymocytes. (H) Quantification of G showing three-dimensional radial position of FISH foci between nuclear periphery (=1.0) and center of the nucleus (=0.0). (I) Proportion of DE genes between Suv39DKO and control cells that overlap disrupted LADs and nondisrupted LADs.











