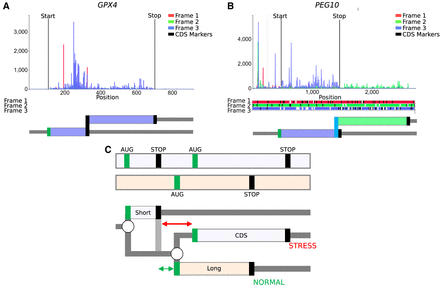
RDG representations of special cases. (A) mRNA encoding selenoprotein GPX4 and (B) PEG10 mRNA requiring ribosomal frameshifting for its expression. Trips-Viz screenshots of ribosomal profiling density for these mRNAs are at the top with ORF plots beneath and RDG representations further below. (C, top) An ORF plot containing three translons (protein-coding CDS and two uORFs, short and long), which is a minimal requirement for the mechanism of delayed reinitiation. (Below) The corresponding RDG with green and red representing the predominant ribosome paths for normal and stress conditions, respectively. Arrows indicate the distance sufficient for scanning ribosomes to bind ternary complexes under normal (green) or stress (red) conditions.











